On automated model home pages
I haven’t written a post in a while! Life, etc. but I’m excited to write about something I’ve thought about on and off again for a bit. If you want to skip the reading and go straight to some code, check out how to automatically document your models in a GitHub repository here.
This post assumes some familiarity with version control and GitHub.
Why a homepage
R and its ecosystem of packages and tooling has made it much easier to statistically model and communicate results. However, one thing that I’ve thought would be useful is a homepage for models. Something that lives close to the code and could automatically help catalog results and diagnostics for a person or a team. I’ve been inspired by the way Hugging Face in particular has been able to host all the information about their uploaded models (mainly for predictive tasks) in a transparent and helpful way for their users.
These homepages wouldn’t replace more involved and customized document generation with R markdown, which I think is wonderful, but would serve as an automatic, transparent and unified interface for cataloging model results.
Leveraging GitHub READMEs
Assuming you use GitHub and your modeling work is version controlled, GitHub already creates “homepages” for your repository using a README.md file. We can enrich a repository README with model results and diagnostics, and have that always be the first thing someone sees when they find your repo.
I created a template repository to automatically do just that!
If you change the code in R/model.R to run your own models, the template will automatically have the repository README update on your pushed changes with documentation for your model results.
How it works
Normally, you create a model with a line of code like lm(Ozone ~ Wind + Solar.R + Temp, data = airquality). If you are using the template, you can simply annotate that line with a comment above,
~ AQ Model
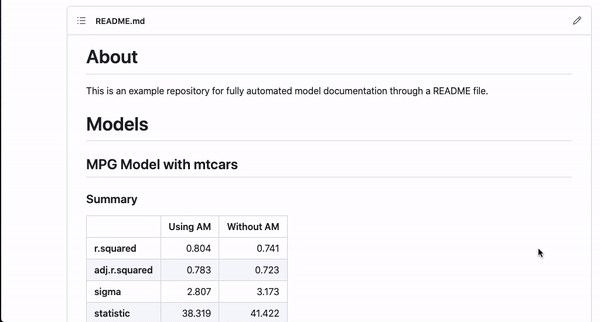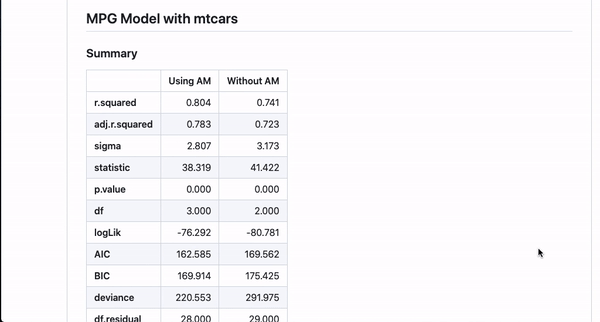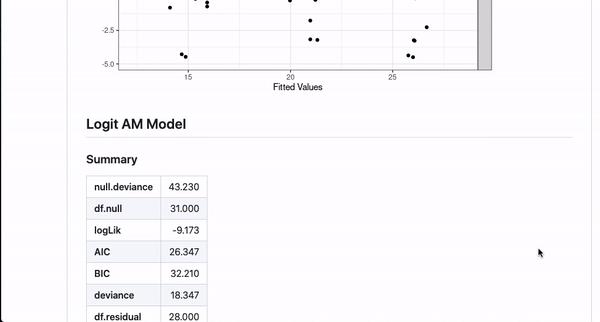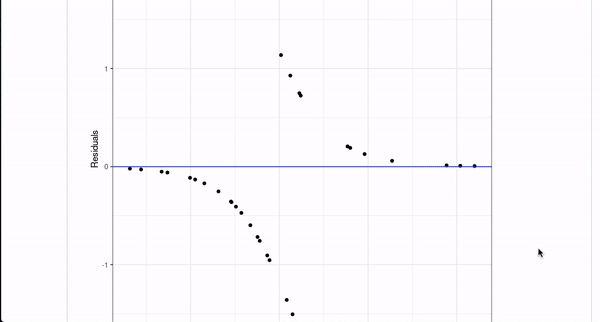
lm(Ozone ~ Wind + Solar.R + Temp, data = airquality)source model_readme/process_file.R and have it add a coefficient table, some overall model summary diagnostics like R^2, and a residuals plot to the README.md.
The #~ indicates to the repository’s code to extract metadata from the model below and the AQ Model is the header that will be added to the README.
All of this is done with the help of the broom package (for working with models in a unified format) and the gt, gtsummary, and ggplot2 packages for displaying data. They are invaluable to making that simple comment create something like:
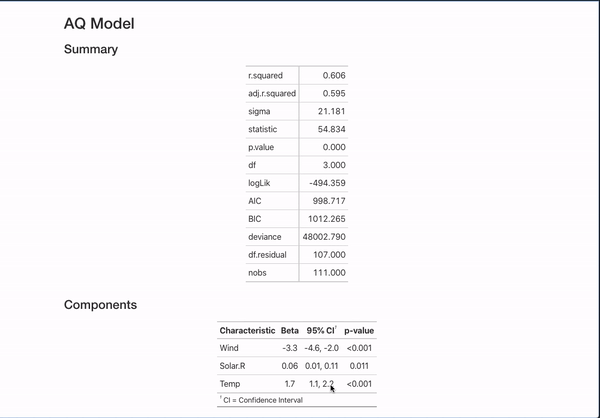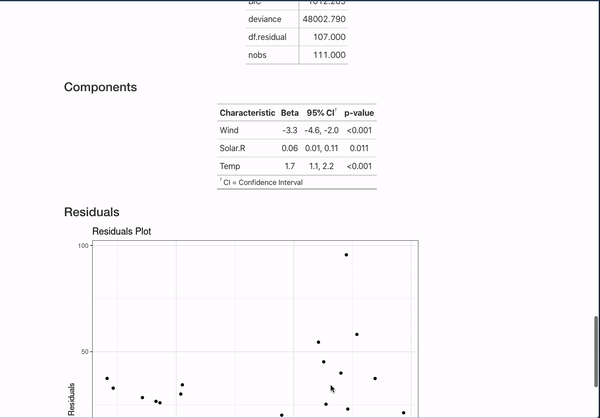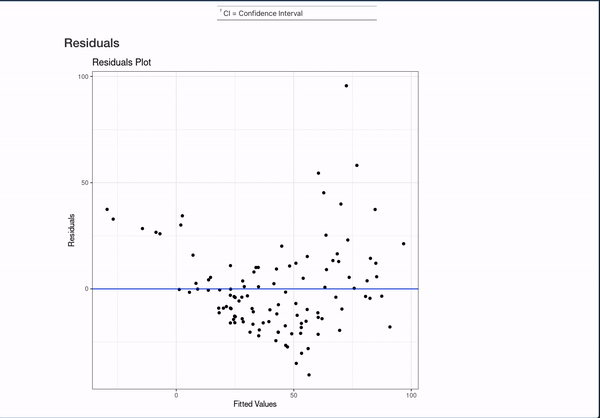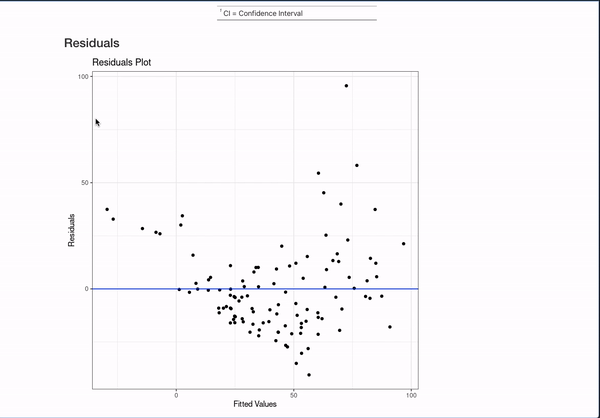
It’s nice to be able to automate this kind of documentation for one model, but a lot of interesting works comes from the comparison of different models studying the same outcome. This template can automate documentation for that as well!
#~ MPG Model with mtcars {Using AM}
m <- lm(mpg ~ cyl + am + hp, data = mtcars)
#~ MPG Model with mtcars {Without AM}
m2 <- lm(mpg ~ cyl + hp,
data = mtcars)If you annotate multiple models with the same header (like here MPG Model with mtcars) and then use {} notation to specifically name the model, the code will automatically combine the models in the README.
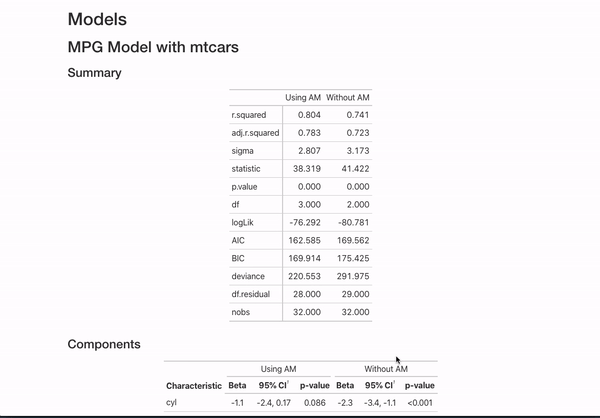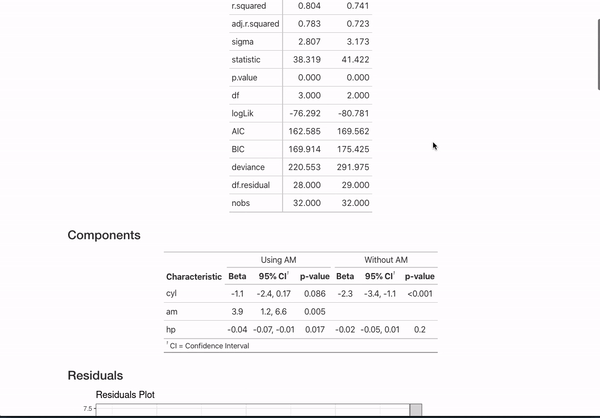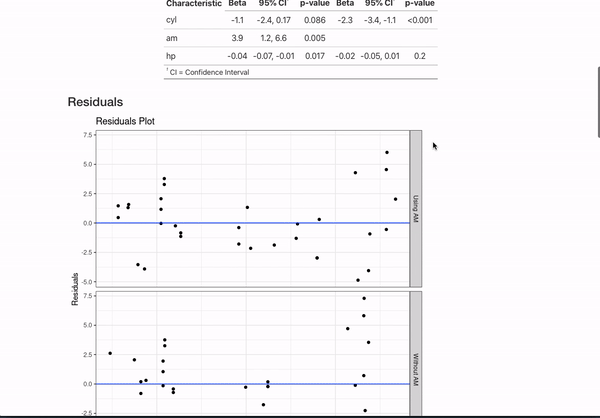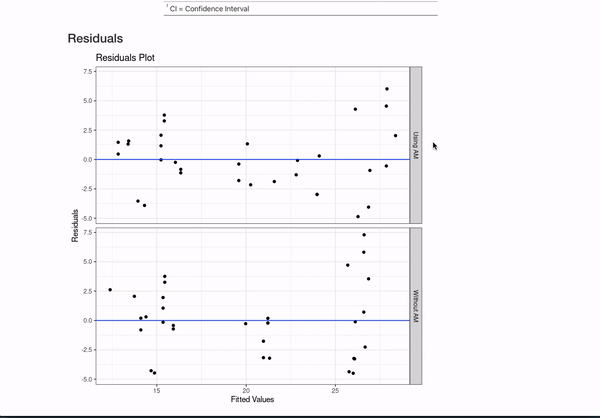
This is all certainly an opinionated set of metadata to display. However, if you have other ideas for customization or want to document your models differently, you can always fork the repo that generates all this documentation and mess around with the functions in model_readme/model_mkdown_outputs.R to get your desired output. And feel free to reach out for any help.
Automating updates
Great so we have a method for creating model homepages. But it is tedious to have to source a file every time you change something. To fully leverage the automated nature of version control, a GitHub Actions workflow was added to fully automate changing the README whenever a commit is pushed to the repository, allowing you to sit back and focus on modeling and let everything else be automated as you make changes.
Make sure you change the repository secret GH_TOKEN to your personal access token if you want to be able to push changes like this automatically.
Future
A lot of this functionality can be wrapped up into an R package. If you think that would be useful, let me know through email or as an issue in the repository where this code is hosted! Hope this is helpful for anyone trying to share their work in an automated way.
Thank you to Dr. William Doane for your help as always.
